How it works
We are recruiting 5,000 participants—people who are doing things to extend life expectancy that most people don’t know about or would not consider. We offer them an extensive questionnaire about everything from social conditions to prescription drugs, and follow up to see which answers change over time. Each person receives a methylation test at the beginning and end of a one-year period.
At the end of the the year, we will make a plot of how much people aged over that time. We expect to see a bell-shaped curve, with most people aging two years, or perhaps a little less if the things we are doing are generally effective.
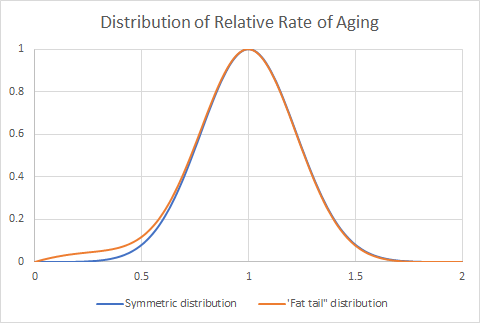
It is the tail ends of that bell curve that has the most information. We will look for a “fat tail” in the bell curve, consisting of an excess of people at the end that suggests very slow aging. We will use data mining and multivariate analysis to find combinations of factors that may account for the especially slow aging achieved by people in this group. If we find factors that are common to this group, then we will have a good candidate for a combination of treatments that is very effective at slowing the aging process. These combinations—if found—will be good candidates for a placebo-controlled trial in the future.
All data will be in the public domain, so that other researchers can pursue other statistical strategies.
More details about the program and the data analysis plan have been published as a peer-reviewed academic article in Rejuvenation Research and in a special aging issue of Biochemistry.